Tumor characteristics analysis of multimodal brain images
|
1.Main Contents
1.1Brief Introduction
Brain tumors are a common tumor-like disease with high morbidity and mortality, which may cause visual impairment, hemiplegia, and even death. The gliomas studied in this paper are the most common primary malignant tumors. It originates from glial cells and is a typical malignant tumor with diffuse, invasive, and blurred borders. MRI images with four modalities (T1, T1c, T2 and Flair) are often used for brain tumor diagnosis, as shown in Figure 1.

Figure 1. Brain tumor MRI images with four modalities.
At present, most of the analysis of medical image data requires manual interpretation by radiologist, which is time-consuming and laborious. Moreover, the final segmentation result depends on the radiologist's experience and subjective decision, even the same radiologist is hard to get reproducible results. Therefore, it is necessary to use CNN automatic segmentation algorithm to segment brain tumors automatically.
1.2Purposes
In the aforementioned methods, the encoder conducts multiple down-sampling to obtain high-level features, which loses the positioning accuracy. Using Dice Loss has somewhat eased the problem of category imbalance, but it did not take the different degrees of category imbalance into consideration. In this paper, we introduced a novel convolutional neural network, namely 3D Dilated-U-Net, by replacing the partial convolutional layer in the Res-U-Net with dilated convolutional layer which could get a larger receptive field. This network reduced the number of pooling layers and improved the positioning accuracy of the decoder. Moreover, we used A-Dice Loss to adapt to category imbalance in our training processing. Experimental results indicated that our proposed 3D Dilated-U-Net could achieve better comprehensive segmentation performance.
|
2.Project research progress
2.1 2.1 Dataset establishment
The dataset used in this paper is mainly from MICCAI BraTS 2017, which contains 169 patients (104 GBM, 65 LGG). Each case consists of four 3D MR images with the same size (156×240×240) and one label, all images are labeled by 1-4 experts following the same protocol, and experienced neuro-radiologists approved their annotations. Annotations comprise the GD-enhancing tumor (ET-label 4), the peritumoural edema (ED-label 2), and the necrotic and non-enhancing tumor (NCR/NET-label 1), after pre-processing cropped to 128×128×128 size as the input of the network, then the network the input size is 4×128×128×128.
The noise processing method used in this paper is to directly remove the 1% highest and lowest gray value pre-processing scheme, and then use the N4ITK method to perform bias field correction on T1 and T2 images. Finally, the data of the four modes are normalized according to the sequence, that is, each image is subtracted from the gray average value of the entire sequence of the mode and divided by the variance.
2.2Construction of integrated hybrid deep learning network
The network we designed is used to handle large 3D input blocks of 128x128x128 voxels. Like the traditional U-Net, our architecture contains a contraction path. When we go deep into the network, it encodes an increasingly abstract input representation, then an expansion path that recombines these representations with shallower features to pinpoint the structure of interest. We refer to the vertical depth (the depth of the U shape) as the network layer, and the higher layer is the lower dimensional representation of the feature dimension with higher spatial resolution. The output in the contraction path is calculated by the encoding module. Similarly, we call the processing block in the expansion path a decoding module. Each encoding module has two 3x3x3 dilated convolution layers with a dilated rate of 1 and a dropout layer (Pdrop = 0.3). The encoding module is connected by a 3x3x3 convolution (stride 2), the image size is reduced to half. We increase the feature map resolution in the expansion path by up-scaling (size 2, stride 2) and then 3x3x3 convolution, which convolves the number of feature maps by half (up-sampling module). After up-sampling, the feature map from the expansion path is connected to the feature map from the contraction channel and then passed to the decoding module. The decoding module comprises a 3x3x3 convolution followed by a 1x1x1 convolution and halve the number of signatures. To improve the stability of the network, the split network is integrated on different layers of the network and combined by element sum to form the final network output.
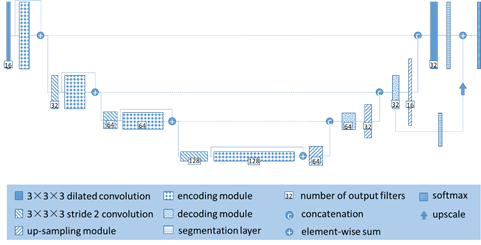
Figure 2. 3D Dilated-U-Net architecture.
|
3.Brain tumor assisted diagnosis system
|
Copyright Announcement
Please respect our original works. Please indicate the original source and author information of the article when reprinting. If you have any questions or ideas and would like to communicate with us, please use the email or phone at the bottom of the page to contact.
|
|